By Aashish Joshi | Founder & CEO

By Aashish Joshi | Founder & CEO
XGBoost belongs to the gradient-boosting class of machine learning models that rely on the decision tree framework. It is arguably one of the most popular machine learning models for regression and classification-based modeling due to its high efficiency. XGBoost model was designed to be used with large, complicated datasets.
For ease of understanding and to better internalize the model, however, the explanation will be based on a dataset with 4 observations, containing 1 independent variable and 1 dependent variable.
Data: The data points below are directly taken from the California Housing Market
dataset –
| AveRooms | MedHouseVal |
| 2.7 | 0.6 |
| 5.5 | 2.8 |
| 6.9 | 4.5 |
| 3.8 | 1.3 |
Here, AveRooms is the average number of rooms per household (independent variable) and
MedHouseVal is the Median Property Valuation in $100,000 (dependent variable)
Implementing the XGBoost Model for Regression:
1. Initial Predictions:
| AveRooms | MedHouseVal | Prediction |
| 2.7 | 0.6 | 0.5 |
| 5.5 | 2.8 | 0.5 |
| 6.9 | 4.5 | 0.5 |
| 3.8 | 1.3 | 0.5 |
2. Calculating Pseudo Residuals:
| AveRooms | MedHouseVal | Prediction | Psuedo Residuals |
| 2.7 | 0.6 | 0.5 | 0.1 |
| 5.5 | 2.8 | 0.5 | 2.3 |
| 6.9 | 4.5 | 0.5 | 4 |
| 3.8 | 1.3 | 0.5 | 0.8 |
3. Building XGBoost Trees:
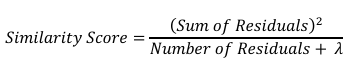
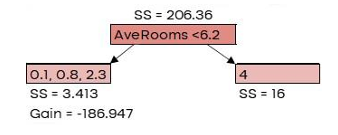
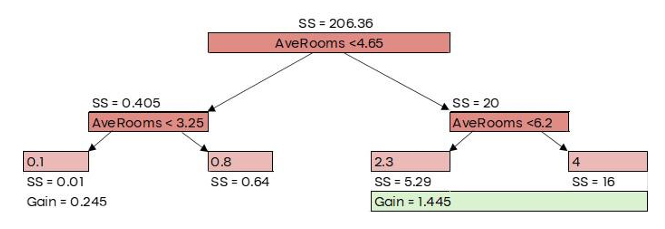
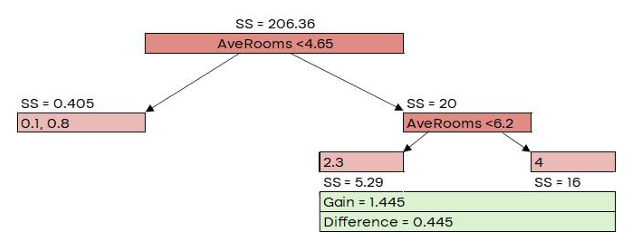
Here, 𝜆 is the regularization parameter, which we’ll set to 0 for now. Similarity Score = 206.36

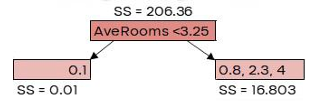
G𝑎𝑖𝑛 = 𝐿𝑒𝑓𝑡 𝑙𝑒𝑎𝑓𝑆𝑆 + 𝑅𝑖𝑔ℎ𝑡 𝑙𝑒𝑎𝑓𝑆𝑆 − 𝑅𝑜𝑜𝑡𝑆�
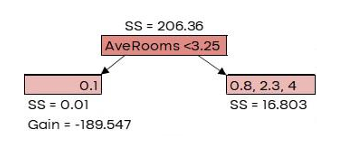
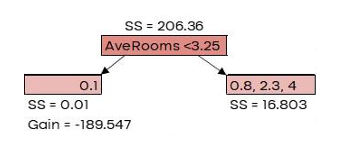
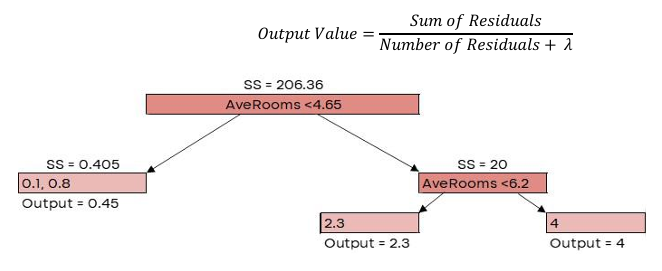
| AveRooms | MedHouseVal | New Prediction |
| 2.7 | 0.6 | 0.635 |
| 5.5 | 2.8 | 1.19 |
| 6.9 | 4.5 | 1.7 |
| 3.8 | 1.3 | 0.635 |
| AveRooms | MedHouseVal | New Predictions |
New Psuedo Residuals |
| 2.7 | 0.6 | 0.635 | -0.035 |
| 5.5 | 2.8 | 1.19 | 1.61 |
| 6.9 | 4.5 | 1.7 | 2.8 |
| 3.8 | 1.3 | 0.635 | 0.665 |
We can see that these new residuals are smaller than the old ones. Hence, we’ve taken a small step in the right direction